DVID is a Distributed, Versioned, Image-oriented Dataservice written to support neural reconstruction, analysis and visualization efforts at HHMI Janelia Research Center. It provides storage with branched versioning of a variety of data necessary for our research including teravoxel-scale image volumes, JSON descriptions of objects, sparse volumes, point annotations with relationships (like synapses), etc.
Its goal is to provide:
- A framework for thinking of distribution and versioning of large-scale scientific data similar to distributed version control systems like git.
- Easily extensible data types (e.g., annotation, keyvalue, and labelmap in figure below) that allow tailoring of APIs, access speeds, and storage space for different kinds of data.
- The ability to use a variety of storage systems via plugin storage engines, currently limited to systems that can be viewed as (preferably ordered) key-value stores.
- A stable science-driven HTTP API that can be implemented either by native DVID data types or by proxying to other services.
How it's different from other forms of versioned data systems:
- DVID handles large-scale data as in billions or more discrete units of data. Once you get to this scale, storing so many files can be difficult on a local file system or impose a lot of load even on shared file systems. Cloud storage is always an option (and available in some DVID backends) but that adds latency and doesn't reduce transfer time of such large numbers of files or data chunks. Database systems (including embedded ones) handle this by consolidating many bits of data into larger files. This can also be described as a sharded data approach.
- All versions are available for queries. There is no checkout to read committed data.
- The high-level science API uses pluggable datatypes. This allows clients to operate on domain-specific data and operations rather than operations on generic files.
- Data can be flexibly assigned to different types of storage, so tera- to peta-scale immutable imaging data can be kept in cloud storage while smaller, frequently mutated label data can be kept on fast local NVMe SSDs. This also allows data to be partitioned across databases by data instance. Our recent datasets primarily hold local data in Badger embedded databases, also written in the Go language.
- (Work in progress) A newer storage backend (DAGStore) will allow "chained storage" such that data published at a particular version, say on AWS Open Data, could be reused for later versions with only new modifications stored locally. This requires extending storage flexibility to versions of data across storage locations. DAGStore will greatly simplify "pull requests" where just the changes within a set of versions are transmitted between separate DVID servers.
While much of the effort has been focused on the needs of the Janelia FlyEM Team, DVID can be used as a general-purpose branched versioning file system that handles billions of files and terabytes of data by creating instances of the keyvalue datatype. Our team uses the keyvalue datatype for branched versioning of JSON, configuration, and other files using the simple key-value HTTP API.
DVID aspires to be a "github for large-scale scientific data" because a variety of interrelated data (like image volume, labels, annotations, skeletons, meshes, and JSON data) can be versioned together. DVID currently handles branched versioning of large-scale data and does not provide domain-specific diff tools to compare data from versions, which would be a necessary step for user-friendly pull requests and truly collaborative data editing.
Users should install DVID from the releases. The main branch of DVID may include breaking changes required by our research work.
Developers should consult the install README where our conda-based process is described.
DVID has been tested on MacOS X, Linux (Fedora 16, CentOS 6, Ubuntu), and Windows Subsystem for Linux (WSL2). It comes out-of-the-box with several embedded key-value databases (Badger, Basho's leveldb) for storage although you can configure other storage backends.
Before launching DVID, you'll have to create a configuration file
describing ports, the types of storage engines, and where the data should be stored.
Both simple and complex sample configuration files are provided in the scripts/distro-files
directory.
Some documentation is available on the DVID wiki for how to start the DVID server. While the wiki's User Guide provides simple console-based toy examples, please note that how our team uses the DVID services is much more complex due to our variety of clients and script-based usage. Please see the neuclease python library for more realistic ways to use DVID at scale and, in particular, for larger image volumes.
Both high-level and detailed descriptions of DVID and its ecosystem can found here:
- A high-level description of Data Management in Connectomics that includes DVID's use in the Janelia FlyEM Team.
- Paper on DVID describing its motivation and architecture, including how versioning works at the key-value level.
- The main place for DVID documentation and information is dvid.io. The DVID Wiki in this github repository will be updated and moved to the website.
DVID is easily extensible by adding custom data types, each of which fulfill a minimal interface (e.g., HTTP request handling), DVID's initial focus is on efficiently handling data essential for Janelia's connectomics research:
- image and 64-bit label 3d volumes, including multiscale support
- 2d images in XY, XZ, YZ, and arbitrary orientation
- multiscale 2d images in XY, XZ, and YZ, similar to quadtrees
- sparse volumes, corresponding to each unique label in a volume, that can be merged or split
- point annotations (e.g., synapse elements) that can be quickly accessed via subvolumes or labels
- label graphs
- regions of interest represented via a coarse subdivision of space using block indices
- 2d and 3d image and label data using Google BrainMaps API and other cloud-based services
Each of the above is handled by built-in data types via a Level 2 REST HTTP API implemented by Go language packages within the datatype directory. When dealing with novel data, we typically use the generic keyvalue datatype and store JSON-encoded or binary data until we understand the desired access patterns and API. When we outgrow the keyvalue type's GET, POST, and DELETE operations, we create a custom datatype package with a specialized HTTP API.
DVID allows you to assign different storage systems to data instances within a single repo, which allows great flexibility in optimizing storage for particular use cases. For example, easily compressed label data can be store in fast, expensive SSDs while larger, immutable grayscale image data can be stored in petabyte-scale read-optimized systems like Google Cloud Storage.
DVID is written in Go and supports pluggable storage backends, a REST HTTP API, and command-line access (likely minimized in near future). Some components written in C, e.g., storage engines like Leveldb and fast codecs like lz4, are embedded or linked as a library.
Command-line and HTTP API documentation can be found in help constants within packages or by visiting the /api/help HTTP endpoint on a running DVID server.
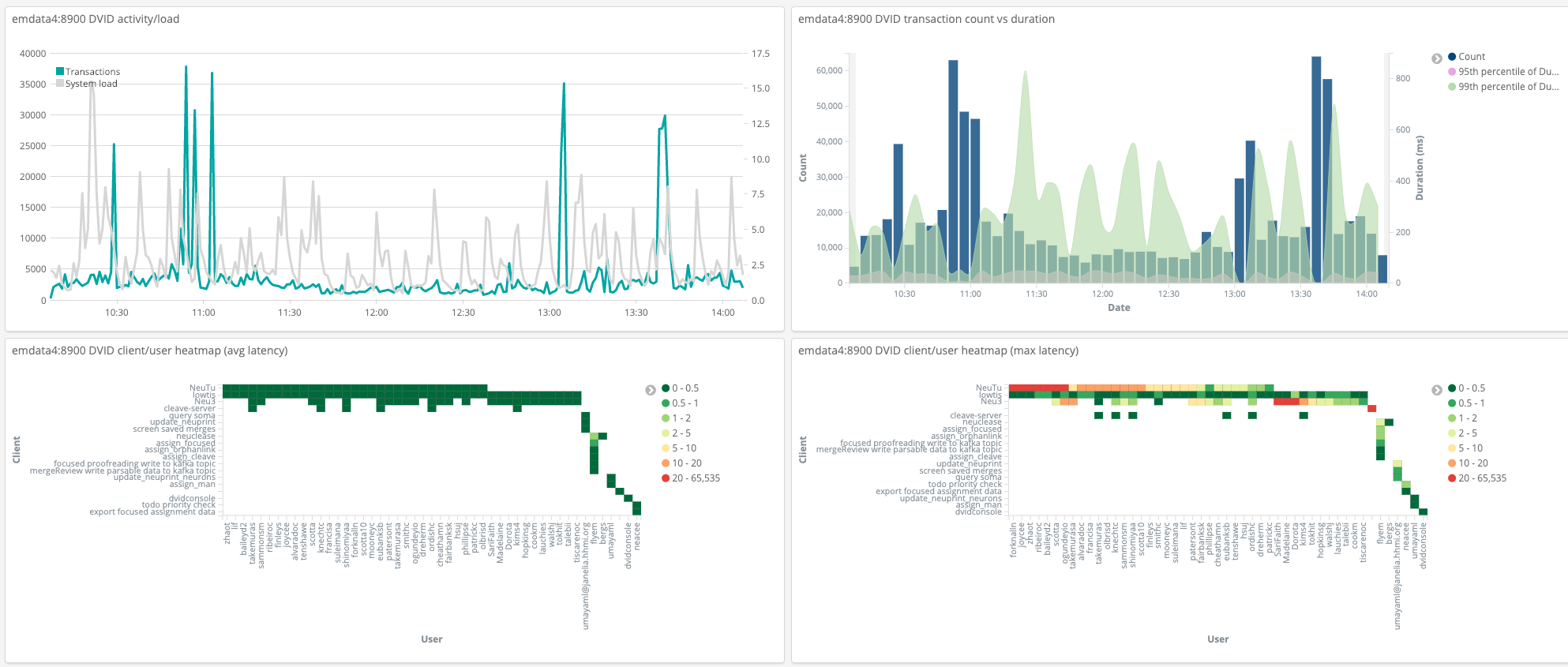
Mutations and activity logging can be sent to a Kafka server. We use kafka activity topics to feed Kibana for analyzing DVID performance.
Programmatic clients:
- neuclease, python library from HHMI Janelia
- intern, python library from Johns Hopkins APL
- natverse, R library from Jefferis Lab
- libdvid-cpp, C++ library from HHMI Janelia FlyEM
GUI clients:
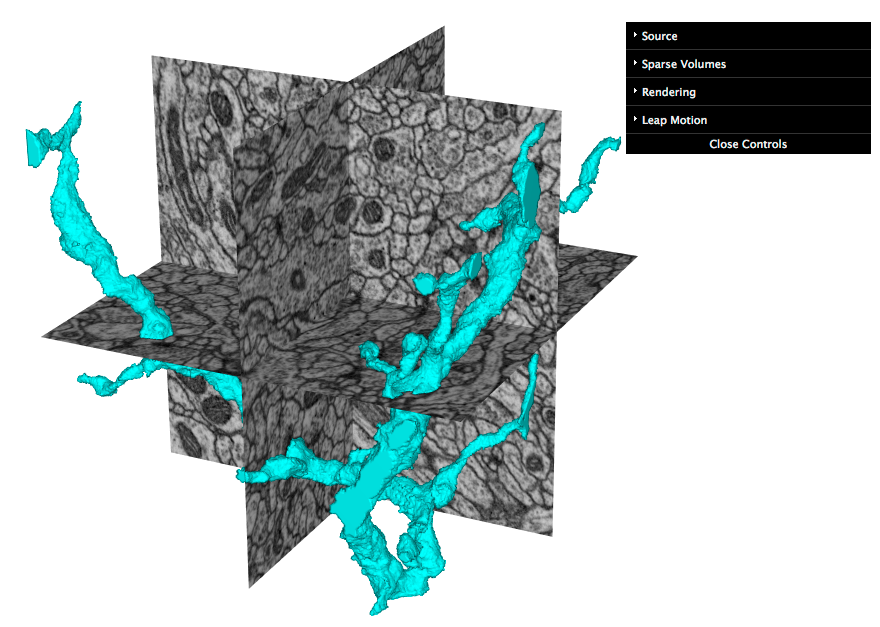
Screenshot of an early web app prototype pulling neuron data and 2d slices from 3d grayscale data: